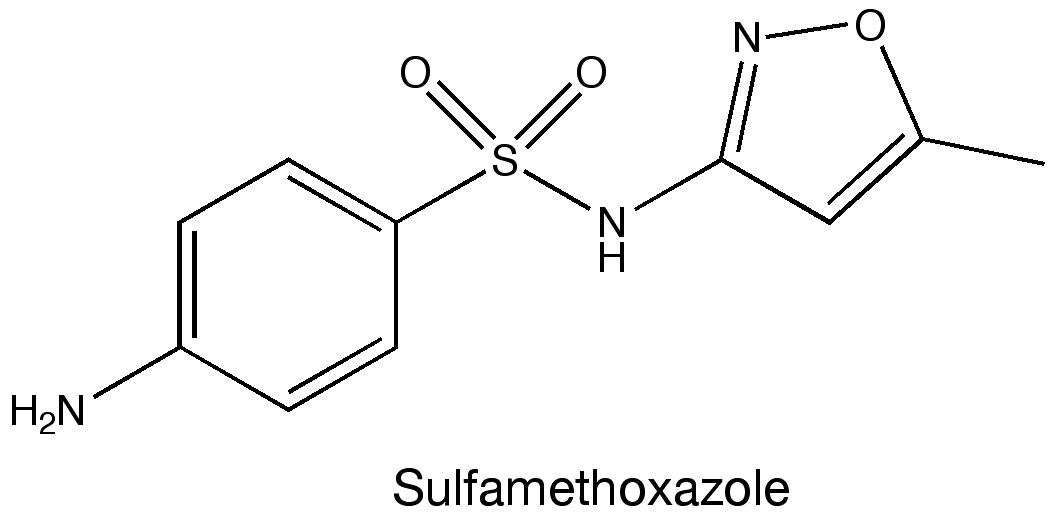
A few years ago during a visit to the hospital, I was infected with a MRSA, or multidrug-resistant Staphylococcus aureus. MRSA refers to any strain of the staphylococcus bacteria that doesn’t respond to treatment with many commonly used antibiotics. Luckily the strain that infected me wasn’t especially virulent; on a return trip the doctor prescribed a sulfamethoxazole & trimethoprim and sent me on my way. The infection cleared soon after.
Sulfamethoxazole (see figure above) belongs to a class of drugs called sulfonamides, or sulfa drugs. These drugs were some of the first antimicrobial treatments and have been used clinically for more than 70 years. So I was intrigued to come across this paper in Science detailing their mechanism of action as I assumed this had been figured out long ago.
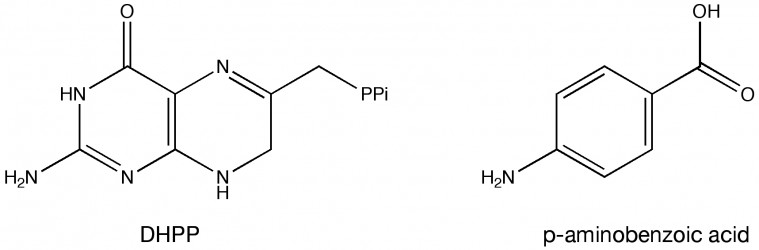
The researchers focused their attention on the enzyme dihydropteroate synthase. This enzyme catalyzes an important reaction in the synthesis of folic acid, which is a B-vitamin microbes need to synthesize their DNA and some amino acids. Using X-ray protein crystallography, they were able to look at the structure of dihydropteroate synthase enzymes from B. anthracis and Y. pests bacteria. To see how the enzyme functions they added two molecules to the protein mixture — 6-hydroxymethyl-7,8-dihydropterin pyrophosphate (DHPP) and para-aminobenzoic acid (see Figure 2). This allowed them to examine the enzyme in its most natural state. The crystal structures with para-aminobenzoic acid and DHPP are bound show that two loops that stabilize intermediates and facilitate product formation. Mutating certain residues in these loops can prevent DHPP or para-aminobenzoic acid from binding.
Later, they added the sulfa drug sulfamethoxazole and they were able to compare how this molecule bound the enzyme versus the two natural ligands. The crystal structures revealed that sulfamethoxazole binds to the protein in a region that overlaps with the binding site of para-aminobenzoic acid. However, sulfamethoxazole extends outside of the binding pocket in such a way that mutating residues in the two loops completely prevents the drug from binding. Mutating the residues Phe33, Thr67, and Pro69 in the loops were specifically mentioned as being linked to sulfa drug resistance. These results provide insight into how microbes become resistant to drugs and also may be able to help identify newer antibiotics.
Reference: M-K Yun et al., “Catalysis and Sulfa Drug Resistance in Dihydropteroate Synthase”. Science (2012), 335, p1110-1113.