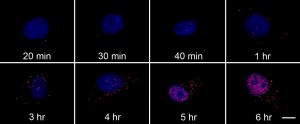
Each red dot is a single probe bound to the RNA target. The cell nucleus is stained blue. Note the increase of fluorescence over time.
Scientists have developed a probe to bind and fluoresce in the presence of a single copy of viral RNA. Up until now, probes fluoresced nonspecifically to other nucleotides limiting their usefulness. A group of scientists use the FISH (fluorescence in situ hybridization) technique to improve things. From CEN:
Yong Chen, at the University of California, Los Angeles, and his colleagues wanted to design a more sensitive assay. They built a long single-stranded DNA containing about 1,000 copies each of two sequences: one complementary to a portion of the RNA that makes up the influenza A virus and another complementary to a fluorescently labeled strand of DNA. They designed the sequences so that they didn’t complement sequences found in their target cells.
To test their new probe, the scientists added it to dog kidney cells infected with influenza A. They had chemically frozen the cells’ biomolecules before adding the probe. After washing away unbound probe, they added the fluorescently labeled strand of DNA.
When the researchers looked at the cells under a microscope, single molecules of viral RNA appeared as bright red dots. They assumed their probes bound a single RNA molecule because the viral nucleic acid concentrations early in the infection were much smaller than that of their DNA probe. The scientists didn’t see the glowing spots in uninfected cells, indicating very little off-target binding, Chen says. His team repeated the experiment with infected cells treated with the antiviral drug ribavirin. Six hours after infection, the amount of viral RNA in those cells was one-twelfth that of untreated cells.
